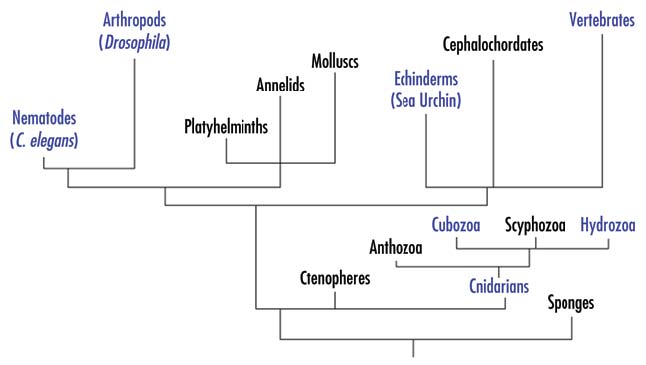
Figure 1. Phylogenetic relationships among various animal groups.
For simplicity, some animal groups have been omitted.
To the table of contents 'Science and Religion'
Correspondence to: Michael Sherman; Department of Biochemistry; Boston University Medical School; 715 Albany St.; Boston, Massachusetts 02118 USA; Tel.: 617.638.5971; Fax: 617.638.5339; Email: sherma1@bu.edu
Original manuscript submitted: 04/05/07
Manuscript accepted: 06/06/07
Previously published online as a Cell Cycle E-publication: http://www.landesbioscience.com/journals/cc/article/4557
I am grateful to Drs. Pinchas Polonsky, Kim Lewis, Slava Epstein, Igor Pechersky and Hirsh Idelson for very helpful discussion.
Recent advances in paleontology, genome analysis, genetics and embryology raise a number of questions about the origin of Animal Kingdom. These questions include: (1) seemingly simultaneous appearance of diverse Metazoan phyla in Cambrian period, (2) similarities of genomes among Metazoan phyla of diverse complexity, (3) seemingly excessive complexity of genomes of lower taxons and (4) similar genetic switches of functionally similar but non-homologous developmental programs. Here I propose an experimentally testable hypothesis of Universal Genome that addresses these questions. According to this model, (a) the Universal Genome that encodes all major developmental programs essential for various phyla of Metazoa emerged in a unicellular or a primitive multicellular organism shortly before the Cambrian period; (b) The Metazoan phyla, all having similar genomes, are nonetheless so distinct because they utilize specific combinations of developmental programs. This model has two major predictions, first that a significant fraction of genetic information in lower taxons must be functionally useless but becomes useful in higher taxons, and second that one should be able to turn on in lower taxons some of the complex latent developmental programs, e.g., a program of eye development or antibody synthesis in sea urchin. An example of natural turning on of a complex latent program in a lower taxon is discussed.
In this essay, I will focus on the origin and evolution of Metazoa, without discussing evolution of unicellular organisms, or other groups. Ideas proposed below in no sense challenge the existing views on formation of small taxonomic units. Our focus, however, will be on emergence of new complex systems, and related appearance of large taxons.
Almost all currently existing Metazoan phyla emerged during a relatively short Cambrian period around 510–550 million years ago (Cambrian Explosion) (reviewed in refs. 1 and 2). In previous periods paleontologists find diverse fauna of unicellular organisms and spongi. Shortly before Cambrian period some Cnidarian and Ediacaran fauna was found, but no other Metazoa. The appearance in evolution of the entire Metazoan fauna seems to have been very sudden. Interestingly, even in early Cambrian layers, in addition to primitive representatives of various phyla, more advanced forms, including relatively complex Crustaceae were discovered.3 Based on these data it was suggested that diversification of Metazoa started way before Cambrian period, however this suggestion appeals to existence of effectively unfossilizable forms, making these types of organism paleontologically “invisible”. This idea is supported by reports of putative trace fossils (e.g., tracks or burrows) dating to pre-Cambrian era. These claims, however, raise a question why fossilizable forms of various phyla appeared almost simultaneously, and were generally refuted, as discussed in recent review (ref. 2). Therefore, it appears that there was no sequential appearance of the major Metazoan taxons from simpler to more complex phyla, as would be predicted by the classical evolutionary model.

Another question comes from studies of genomes of existing phyla. It appears that emergence of multicellular animals coincided with a dramatic increase in genome complexity in terms of both gene number and appearance of entirely novel gene families. In fact, in unicellular eukaryotic organisms, like yeast, the gene number is usually in the range of five to eight thousand (although in a recently published Tetrahymena genome there is about 28,000 genes, this large number of genes is achieved simply by multiple duplications). On the other hand, in a relatively primitive roundworm C. elegans there is around 19,000 genes (these and following data on the genomes of different species are taken from the corresponding Genome Projects). Similarly, the number of genes in representatives of Cnidaria is around 20,000. Further evolution towards humans did not generate principally bigger genomes, since the sea urchin has around 24,000 genes, and humans possess somewhat below 25,000 genes. Moreover, some taxons show a clear trend of reduction in the number of genes as the complexity increases. For example, the number of genes in the fruit fly Drosophila appears to be reduced to less than 14,000 in spite of an enormous increase in morphological and behavioral complexity compared to a medusa or roundworm, with many genes found in worms missing in Drosophila. These findings suggest that there is excessive genetic information in corals or roundworms, and possibly other taxons as well. Below we will discuss the excessive genetic information in more details.
Among entirely novel families of genes that appeared in Metazoa are genes associated with formation of body plans, organ development and cell‑cell communications, for example Hox, TGFb, Wnt, nuclear receptor systems, etc. Some of them are clearly involved in complex developmental programs, associated with bilateral organization. Existing data from morphology suggest that bilateral taxons emerge from organisms with radial symmetry, like Cnidaria. Whether this suggestion is correct or these groups evolved independently, one does not expect to find genes responsible for development of bilateral organisms in primitive Metazoa with radial symmetry. Surprisingly, such genes, e.g., orthologs of hox genes, were found in Cnidaria, and furthermore they are expressed in Cnidaria in an asymmetric manner, as if to define segments in these radial organisms.6 As noted in the original publication, the overall complexity of regulatory genes and signaling pathways in Cnidaria “is paradoxical, given that this organism contains apparently few tissue types and the simplest extant nervous system consisting of a morphologically homogenous nerve net”.4 For example, the regulatory genes existing in the coral genus Acropora include five TGFb family members, representing four out of six subfamilies found in vertebrates, and downstream components of the signaling pathways; representatives of all Smad subfamilies; a large variety of multiple components of Ras MAPK pathways, etc.5, 7 The three‑way comparisons show that most ESTs of Acropoda match human sequences much more strongly than they do any Drosophila or C. elegans sequence.4, 9 As noted in an original publication, “in many respects, the complexity of the Anthozoan (class of Cnidaria) gene set does not differ substantially from that of vertebrates, and frequently exceeds that of the model invertebrates Drosophila and Caenorhabditis”.5 In fact, many of the regulatory genes were lost later in evolution, and are not present in Drosophila or C. elegans, e.g., hedgehog gene,5 indicating that their presence is not necessary for development and life of very complex Arthropoda. A recently sequenced genome of sea urchin (see Fig. 1) represents another very clear example of a seemingly excessive genetic complexity. As mentioned above, the relatively simple sea urchin has about 24,000 genes, same as more complex vertebrates.9 Though sea urchin lacks eyes and, of course, brain, it has six opsins, belonging to several families found in humans, Drosophila, Scallops and other groups.10 While the presence of the opsins could be explained by their possible function in a simple light sensing, sea urchin has the entire set of orthologs of major genes involved in the eye development, e.g., Pax6, Six3, Prox1, Rx2 or Eya1 (NCBI database). Therefore, it appears that information on the eye development is encoded in the sea urchin genome, while no eye is actually developed, and thus the genetic information seems to be excessive.
Another surprise came from a complexity of components of the immune system in sea urchin. In addition to an extremely well developed system of the innate immunity, these animals possess genes encoding major components of the adaptive immune response. For example, several families of immunoglobulin domain genes (a total of about fifty) have been identified that are predicted to encode immunoglobin variable‑type (V) domains similar to those used by adaptive immune receptors of vertebrates.11 Also, sea urchin has Rag1 and Rag2 genes that mediate the somatic rearrangement process common to both immunoglobulin and T cell‑antigen receptor gene families. In addition, other components that function in the reorganization and diversification of immunoglobulins and TCR have also been identified, including a polymerase homologous to the terminal deoxynucleotidyl transferase (TdT) and polymerase m.11 Yet, sea urchin does not have antibodies, and possibly lacks adaptive immunity in general. Genes that are seemingly useless in sea urchin but are very useful in higher taxons exemplify excessive genetic information in lower taxons.
It is unclear how such genetic complexity could have evolved. In fact, the seemingly excessive complexity probably cannot be explained within the classical model in principle, since this model requires a direct link between acquiring of novel genetic information and its functional importance for improved fitness. A possible response to these arguments within the classical model would be a suggestion that the genes responsible for eye development in Arthropoda or vertebrates serve different functions in lower taxons (so‑called gene sharing). In fact, several examples of gene sharing have been described, e.g., recruiting of small heat shock proteins to serve as crystallines. These examples, however, are exceptionally rare, and it is unclear whether they indeed can be responsible for making de‑novo complex developmental programs from genes serving unrelated functions. The plausibility of this hypothesis at the very minimum is questionable, and must require an experimental prove. On the other hand, the excessiveness of the genetic information in lower taxons is a testable possibility that will be discussed below.
A distinct set of data that call for a novel approach to evolution comes from comparison of genes that control functionally similar genetic programs in Chordata and Arthropoda. There appears to be a high degree of similarity in some of these genes. A classic example of such similarity is Pax6 gene that controls development of visual systems. According to all current accounts, a common ancestor of Chordata and Arthropoda was a very primitive organism that lacked eyes, and therefore the evolution of eyes in these groups was convergent. Nevertheless, Pax6 gene controls eye development in both groups, and Pax6 from Drosophila is replaceable with the mouse gene.12 The same is true about other genes controlling eye development. Moreover, the eye development is not unique in that sense, since the development of certain other functionally similar organ systems or behavioral programs that were not present in a common ancestor are also controlled by similar master switch genes (for example, control of speech development in humans and singing in birds by the FoxP2 gene, ref. 13). So, how does it happen that convergently evolved systems have same developmental switches? These findings are very difficult to explain within the context of Darwinian ideas.
Therefore, we need to explain the following facts: (1) seemingly simultaneous appearance of paleontological remains of all presently existing Metazoan phyla, both simple and advanced; (2) similarities of genomes among Metazoan phyla of diverse complexity; (3) seemingly excessive complexity of genomes of lower taxons; (4) similar genetic switches of functionally similar but non-homologous developmental programs.
Of note, in thinking about metazoan evolution, one should realize that any evolutionary event represents changes in developmental programs, rather than changes in a developed organism. For example, appearance of hooves in horses reflects a change in the program of limb development. Therefore, we need to think about biological evolution in terms of evolution of developmental programs.
Here I propose a hypothesis that answers the questions posed above, and offers experimentally testable predictions. This hypothesis postulates that (1) shortly (in geological terms) before Cambrian period a Universal Genome that encodes all major developmental programs essential for every phylum of Metazoa emerged in a unicellular or a primitive multicellular organism; (2) The Metazoan phyla, all having similar genomes, are nonetheless so distinct because they utilize specific combinations of developmental programs. In other words, in spite of a high similarity of the genomes in phyla X and Y, an organism belonging to phylum X expresses a specific set of active developmental programs, while an organism belonging to a different phylum Y has a distinct set of “working” programs specific for phyla Y. This seemingly trivial statement changes the whole perception of evolution, claiming that the placement of an organism to a particular taxon depends on expression of a specific set of pre-existing developmental programs, rather than on difference in the genetic information per se. Therefore, within the Universal Genome model, what we perceive as a sequential evolution is actually a reflection of expression of one or another combination of programs from the Universal Genome. These postulates explain a simultaneous emergence of Metazoan phyla during Cambrian period, as well as similarities of genomes and a dramatic increase in genome complexity in Metazoan phyla. They also explain similarities of the general switches of functionally similar, but evolutionary distinct developmental programs.
The proposed model of Universal genome implies that a lot of information encoded in genomes is not utilized in each individual taxon, and therefore is effectively useless. Thus, within this model it is not possible in principle to assign functionality to every ORF and its importance for fitness.
The “Universal Genome” hypothesis does not contradict any well‑established data on the genetic evolution (e.g., gene duplications or accumulation of mutations, molecular clock, etc), but suggests that genetic evolution could shape and improve function of developmental programs. Furthermore, genetic evolution in combination with natural selection could define microevolution, however, within this model it is not responsible for the emergence of the major developmental programs.
Before discussing ways to test the proposed model, let’s consider a fundamental problem that the model poses—if there is a Universal Genome, why many genes that are found in higher taxons are not found in lower taxons. This apparent problem could be explained not by a progressive evolution of more complex gene systems from primitive forms, but rather by the loss of certain unused elements of the Universal Genome in primitive forms during the last 530 million years. Loss of genetic information in evolution of complex taxons has been mentioned previously, i.e., much fewer genes in Drosophila compared to C. elegans. A beautiful illustration of such a loss is a recent analysis of Wnt gene family. In humans, there are nineteen Wnt genes belonging to twelve families.14 In Hydra, on the other hand, there are two Wnt genes that correspond to two families found in humans.15 Simple analysis of this finding within the framework of the classical model suggests that additional human genes have developed from ancestral Wnt genes found in Hydra. However, Anemona that belongs to a distinct branch of Cnidaria has eleven Wnt genes belonging to eleven families found in humans.14 Therefore, it is quite obvious that many Wnt gene families, possibly the entire set, exists in the gene pool of the primitive Metazoan phylum, and various members of Wnt families were lost in different species within this phylum. Accordingly, the proposed model predicts that in various groups of Cnidaria we will find many diverse gene families that function in more advanced phyla. Of course, a degree of homology between human genes and related genes in Cnidaria could be masked due to mutations, which must especially affect Cnidaria genes that are not in use.
Since Drosophila lost a lot of genes, which are retained in the Cnidaria genome, according to our model there may be special mechanisms of gene conservation in Cnidaria and possibly other lower taxons, which are not functional in more developed Arthropoda. These mechanisms could be analogous to mechanisms of supporting identical multiple copies of ribosomal RNA genes.
In line with our model, molecular evolution trees often do not fit a morphology‑based evolution tree. For example, there are several TRAF genes in humans and Drosophila, and obvious prediction of Darwin’s model is that there must be an ancestral gene in a common ancestral organism from which the modern TRAF genes were derived. In reality, however, a TRAF gene from the hydroid Hydractinia does not fit criteria of an ancestral gene, which must be somewhat of a mix of all human TRAF families, but rather clearly belongs to the major group of TRAF genes along with human TRAF1, TRAF2, TRAF3 and TRAF 5, while human TRAF4 and especially TRAF6 belong to different groups together with Drosophila TRAFs.16 Such a contradiction with the Darwinian model could be easily explained within the proposed model, according to which all major groups of TRAFs existed within the Universal Genome, and Hydractinia lost many of them, while humans retain them. On the other hand, some of the new members of the TRAF1‑TRAF5 group could be derived from the ancestral Hydra‑TRAF‑like gene via duplications.
There are two main testable predictions of the presented hypothesis, which are absolutely critical for validation of the model: (1) full or parts of the developmental programs characteristic to higher taxons must be encoded in genomes of lower taxons, and (2) blocks of genetic information encoding these developmental programs in more primitive taxons must be useless in these taxons.
Interestingly, Nature appears to provide us with an example of existence of a complex developmental program that suddenly emerges in a primitive organism and acquires full functionality in distant higher taxons. As mentioned above, the common ancestor of Arthropoda and Chordata most likely was a primitive organism without eyes. Nevertheless, Cubozoan jellyfish has well‑developed eyes as a part of a special sensory organ ropalium.17 Each of the four rhopalia in medusa has six separate light‑sensing structures, and a statocyst, a mechanosensory organ and putative ancestor of the inner ear. There are two complex, lens‑containing camera eyes (a bigger and smaller) situated at right angles to each other, and two pairs of simple ocelli comprising photoreceptors (ref. 18). The camera‑type eyes of Cubozoa are much alike that of the squid or even vertebrate eyes, and are way more complex than primitive eyes of other jellyfish. All major components of a typical camera‑type eye are present, including cornea, lens, retina, pigment layer and iris.17 The lenses are spherical. The lower eye has a mobile pupil that can close down the aperture, whereas the pupil of the upper eye remains constant at all light intensities. Light path analysis showed that the upper‑eye lens produces an almost aberration‑free focus.19 Therefore, it can produce sharp images. Most suprizingly, the focal distance of the lens was much higher than the distance to retina.19 Therefore, the visual resolution of these eyes is much poorer than the sophisticated lens would allow. To explain these peculiarities of the jellyfish eye, it was suggested that there might be visual tasks of medusa that are best served by a blurred image. For example, they may need to be able to overlook small objects, like plankton in order to detect motion of bigger objects, like fish.20 However, this suggestion does not explain why the lenses have a capability to produce sharp images, and how these lenses could have evolved by selection pressure. Another interesting feature of the jellyfish vision system is that the neural network in Cubozoa is organized into sub‑umbrellar rings and ganglia, but there is no central processing unit that we could consider a brain.17,18 Therefore it is unclear whether the visual information about shapes can be processed and integrated.
Although Cubozoa do not have a straight homolog of Pax6 that controls the eye development in vertebrates and Arthropoda, they have a PaxB gene that appears to be an ancestral gene for Pax6 and Pax2, a gene controlling the development of an ear.17 In a sense, Pax6 represents PaxB that lost its octapeptide domain but retains the homeobox domain, while Pax2 represents PaxB that lost homeobox but retains the octapeptide domain. The close relation between PaxB and Pax6 genes is supported by the fact that ectopic expression of PaxB causes eye development in Drosophila.17 Both morphological and genetic data strongly indicate similarity of programs of eye development in Cnidaria and vertebrates, rather than convergent evolution. Therefore, the presence of an eye in Cnidaria poses the following evolutionary questions that must be explained: (a) appearance of a camera‑type eye in Cnidaria that is morphologically similar to a vertebrate eye, while such organ is absent from a putative common ancestor of Chordata and Arthropoda; (b) similar mechanisms of molecular regulation of eye developmental program in vertebrate, Cnidaria and Arthropoda; (c) development in evolution of the Cnidarian eye that works not to its full capacity (under‑ resolution); (d) lack of brain in Cnidaria, and therefore possibly lack of a way to process the visual information from the four separate ropalia. While it is not clear how these questions can be answered within the Darwinian model, explanation within the proposed model is quite obvious. The program of eye development has been present in the Universal Genome, and is latent in the majority of Cnidaria, and actually other, even more developed animal groups. This program however was turned on in Cubozoa, and led to the eye development in this group.
The program is poorly tuned resulting in an under‑focused eye, suggesting that the Universal Genome contains relatively rough programs, and tuning the programs could occur through Darwinian mutation‑selection mechanisms. The under‑focused eye that by a mutation was activated in Cubozoa may be useless. On the other hand, in the process of further evolution Cubozoa could develop a capability to utilize this organ to some extend, which may be helpful in their relatively complex swimming behavior, compare to other medusa. Lack of a brain however could preclude them from developing an eye that functions at its full focus capacity.
In addition to the example from Nature, experimental testing of the idea that genomes of lower taxons possess information about developmental programs characteristic of higher taxons is absolutely essential for validation of the proposed model. Manipulating transcription factors that regulate developmental programs, or treatments with teratogenes can possibly activate these developmental programs experimentally.
As a general approach to searching for such latent developmental programs one can effectively utilize information from genome projects. Accordingly, one may identify genes responsible for an un‑expressed developmental program in a genome of an organism belonging to a lower taxon, and try to activate it. For example, as mentioned above, the sea urchin genome encodes a large set of genes involved in the eye development. It is possible that overproduction of the active master switch Pax6 in sea urchin may initiate eye development. Similarly, since major components of the adaptive immunity are encoded in the sea urchin genome, one could attempt to activate production of antibodies by experimenting with transcription factors or teratogenes.
Another indication that latent developmental program is present in a lower taxon would be expression of such a program in higher taxons derived from the lower one in a seemingly convergent processes. For example, a possible experiment would be to activate development of circulation systems of mammalian or bird types in lizards, or even in Xenopus. The circulation systems in mammals and birds appear to be very similar, however, they developed from the Reptilian system independently in these taxons. Therefore, it seems likely that Reptilia possess the program of development of the circulation system of the mammalian/bird type and requires only a minor switch to activate it.
The second prediction is that large blocks of information about developmental programs encoded in genomes of lower taxons must be useless. Therefore, deletion of these blocks should not affect physiology of these organisms. For example, deletion of the entire program of development of the adaptive immune response must not affect physiology of sea urchin. Similarly, deletion of eye‑specific developmental regulators like Rx2 must not be critical for the sea urchin physiology.
The success of this or analogous experiments would provide strong support for the hypothesis of Universal Genome.
1. Valentine JW, Jablonsky D. Morphological and developmental macroevolution: A paleon tological perspective. Int J Dev Biol 2003; 47:517‑22.
2. Conway‑Morris S. The Cambrian “explosion” of metazoans and molecular biology: Would Darwin be satisfied? Int J Dev Biol 2003; 47:505‑15.
3. Budd GE, Butterfield NJ. Crustaceans and the cambrian explosion. Science 2001; 294:2047a.
4. Kortschak DR, Samuel G, Saint R, Miller DJ. EST Analysis of the Cnidarian Acropora millepora reveals extensive gene loss and rapid sequence divergence in the model invertebrates. Current Biology 2003; 13:2190‑5.
5. Technau1 U, Rudd S, Maxwell P, Gordon PMK, Saina M, Grasso LC, Hayward DC, Sensen CW, Saint R, Holstein TW, Ball EE, Miller DJ. Maintenance of ancestral complexity and non-metazoan genes in two basal cnidarians. TRENDS in Genetics 2005; 21:633‑9.
6. Matus DO, Pang K, Marlow H, Dunn CW, Thomsen GH, Martindale MQ. Molecular evidence for deep evolutionary roots of bilaterality in animal development. PNAS 2006; 103:11195‑200.
7. Samuel G, Miller D, Saint R. Conservation of a DPP/BMP signaling pathway in the nonbilateral cnidarian Acropora millepora. Evol Dev 2001; 3:241‑50.
8. Ball EE, Hayward DC, Reece‑Hoyes JS, Hislop NR, Samuel G, Saint R, Harrison PL, Miller DJ. Coral development: From classical embryology to molecular control. Int J Dev Biol 2002; 46:671‑8.
9. Sodergren E, Weinstock GM, Davidson EH, et al. Sea urchin genome sequencing consortium. The genome of the sea urchin Strongylocentrotus purpuratus. Science 2006; 314:941‑52.
10 Raible F, Tessmar‑Raible K, Arboleda E, Kaller T, Bork P, Arendt D, Arnone MI. Opsins and clusters of sensory G‑protein‑coupled receptors in the sea urchin genome. Dev Biol 2006; 300:461‑75.
11. Rast JP, Smith LC, Loza‑Coll M, Hibino T, Litman GW. Genomic insights into the immune system of the sea urchin. Science 2006; 314:952‑6.
12. Oliver G, Gruss P. Current views on eye development. Trends Neurosci 1997; 20:415‑21.
13. White SA, Fisher SE, Geschwind DH, Scharff C, Holy TE. Singing mice, songbirds, and more: Models for FOXP2 function and dysfunction in human speech and language. J Neurosci 2006; 26:10376‑9.
14. Kusserow A, Pang K, Sturm C, Hrouda M, Lentfer J, Schmidt HA, Technau U, von Haeseler A, Hobmayer B, Martindale MQ, Holstein TW. Unexpected complexity of the Wnt gene family in a sea anemone. Nature 2005; 433:156-60.
15. Hobmayer B, Rentzsch F, Kuhn K, Happel CM, von Laue CC, Snyder P, Rothbacher U, Holstein TW. WNT signaling molecules act in axis formation in the diploblastic metazoan Hydra. Nature 2000; 407:186‑9.
16. Mali B, Frank U. Hydroid TNF‑receptor‑associated factor (TRAF) and its splice variant: A role in development. Mol Immunol 2004; 41:377‑84.
17. Piatigorsky J, Kozmik Z. Cubozoan jellyfish: An Evo/Devo model for eyes and other sensory systems. Int J Dev Biol 2004; 48:719‑29.
18. Coates MM. Visual ecology and functional morphology of cubuzoa (cnidaria). Integr Comp Biol 2003; 43:542‑54.
19. Nilsson DE, Gislen L, Coates MM, Skogh C, Garm A. Advanced optics in a jellyfish eye. Nature 2005; 435:201‑5.
20. Wehner R. Sensory physiology: Brainless eyes. Nature 2005; 435:157‑9.
www.landesbioscience.com Cell Cycle 1873
